Research projects
- Ⅰ.Biomedical engineering: hydrogel research
- Ⅱ. Research for SARS-CoV-2
- Ⅲ. Basic cancer research: Oncogene and molecular mechanism of cancer progression
- Ⅳ. Brain tumor research
- Ⅴ. Surgical pathology
- Ⅵ. AI and digital pathology
- Ⅶ. Other collaborative researches
Ⅰ.Biomedical engineering: hydrogel research
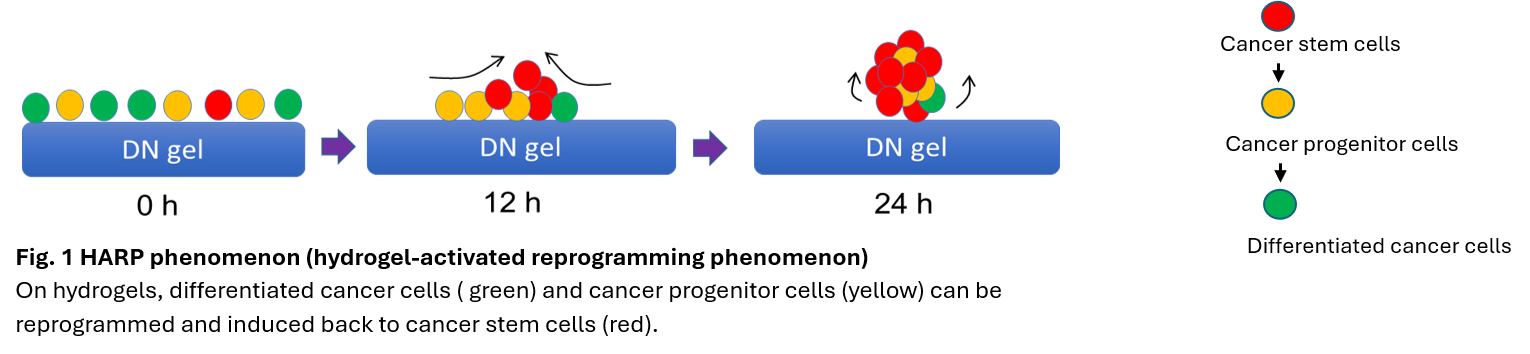
Dr. Tanaka has found that polymer hydrogels with a double-network structure induce cancer cell reprogramming in an extremely short period of time to create cancer stem cells (Fig. 1) (hydrogel-activated reprogramming phenomenon, HARP phenomenon). This is different from conventional cancer stem cell isolation and identification methods, and is a breakthrough method that can identify cancer stem cells within 24 hours regardless of the type of cancer (Nat Biomed Eng, 5, 941-925, 2021). genome. Based on the HARP phenomenon, this study aims to create highly functional gels by optimizing the physical factors of gel monomers, and by integrating multiple spatial information analysis techniques such as the latest single cell spatial transcriptome analysis and Raman microscopy, to elucidate the biological phenomena of cancer and develop therapeutic agents for cancer stem cells, which are the source of relapse. Development of therapeutic drugs for cancer stem cells, which are the source of recurrence. This process of controlling genomes using hydrogels will be created as a new academic field called “Material Genomics”.
Tanaka Lab is focusing on several medical researches including cancer research and regenerative medicine by using hydrogel. Cancer is the leading cause of death in the nation and there is a need to develop curative therapies. Although current therapies reduce or eliminate cancer temporarily, cancer recurs after a few years. The cause of recurrence is the survival of therapy-resistant cancer stem cells in cancer tissue. Therefore, eradication of cancer stem cells would dramatically improve the prognosis. However, cancer stem cells are difficult to detect because of their extremely low abundance, and there are currently no therapies that target these cells. For regenerative medicine, hydrogels are applied to culture iPS cell-derived organoid, neuronal cells, and intestinal organoids. Uncovering the molecular mechanism of HARP phenomenon is another interesting projects in Tanaka lab.
Publications for hydrogel research
Original publications
- Hashimoto D, Semba S, Tsuda M, Kurokawa T, Kitamura N, Yasuda K, Gong J P, and Tanaka S. Integrin α4 mediates ATDC5 cell adhesion to negatively charged synthetic polymer hydrogel leading to chondrogenic differentiation. Biochem Biophys Res Commun, 528, 120-126, 2020.
- Semba S, Kitamura N, Tsuda M, Goto K, Kurono S, Ohmiya Y, Kurokawa T, Gong J P, Yasuda K, and Tanaka S. Synthetic PAMPS gel induces chondrogenic differentiation of ATDC5 cells via a novel protein reservoir function. J Biomed Mater Res A, 1-11, 2020.
- Suzuka J, Tsuda M, Wang L, Kohsaka S, Kishida K, Semba S, Sugino H, Aburatani S, Frauenlob M, Kurokawa T, Kojima S, Ueno T, Ohmiya Y, Mano H, Yasuda K, Gong J P, and Tanaka S. Rapid reprogramming of tumour cells into cancer stem cells on double-network hydrogels. Nat Biomed Eng, 5, 914-925, 2021.
- Ferdous Z, Clément J E, Gong J P, Tanaka S, Komatsuzaki T, and Tsuda M. Geometrical analysis identified morphological features of hydrogel-induced cancer stem cells in synovial sarcoma model cells. Biochem Biophys Res Commun, 642, 41-49, 2023.
- Tanikawa S, Ebisu Y, Sedlačík T, Semba S, Nonoyama T, Kurokawa T, Hirota A, Takahashi T, Yamaguchi K, Imajo M, Kato H, Nishimura T, Tanei Z I, Tsuda M, Nemoto T, Gong J P, and Tanaka S. Engineering of an electrically charged hydrogel implanted into a traumatic brain injury model for stepwise neuronal tissue reconstruction. Sci Rep, 13, 2233, 2023.
- Kato-Shinomiya M, Sugino H, Wang L, Saito Y, He J, Tanei Z I, Oda Y, Tanikawa S, Tanino M, Gong J P, Tsuda M, and Tanaka S. SLC13A5 plays an essential role in the energy shift to oxidative phosphorylation in cisplatin-resistant mesothelioma stem cells. Pathol Int, 75, 151-165, 2025.
- Nie Y, Mu Q, Sun Y, Ferdous Z, Wang L, Chen C, Nakajima T, Gong J P, Tanaka S, and Tsuda M. Mechanochemistry-Induced Universal Hydrogel Surface Modification for Orientation and Enhanced Differentiation of Skeletal Muscle Myoblasts. ACS Appl Bio Mater, 2025.
- Sun Y, Nie Y, Wang L, Gong J P, Tanaka S, and Tsuda M. Tumor-mimetic hydrogel stiffness regulates cancer stemness properties in H-Ras-transformed cancer model cells. Biochem Biophys Res Commun, 743, 151163, 2025.
- Sawai S, Oda Y, Saito Y, Kuwabara T, Wang L, Tanei Z I, Hirabayashi S, Tsuda M, Gong J P, Manabe A, and Tanaka S. Analysis of synthetic polymer hydrogel-based generation of leukemia stem cells. Biochem Biophys Res Commun, 744, 151149, 2025.
- Aoki Y, Wang L, Tsuda M, Saito Y, Kubota T, Oda Y, Hirano S, Gong J P, and Tanaka S. Hydrogel PCDME creates pancreatic cancer stem cells in OXPHOS metabolic state with TXNIP elevation. Biochem Biophys Res Commun, 751, 151416, 2025.
Collaborative publications
- Goto K, Kimura T, Kitamura N, Semba S, Ohmiya Y, Aburatani S, Matsukura S, Tsuda M, Kurokawa T, Gong J P, Tanaka S, and Yasuda K. Synthetic PAMPS gel activates BMP/Smad signaling pathway in ATDC5 Cells, which plays a significant role in the gel-induced chondrogenic differentiation. J Biomed Mater Res A, 734-746, 2015.
- Frauenlob M, King D R, Guo H, Ishihara S, Tsuda M, Kurokawa T, Haga H, Tanaka S, and Gong J P. Modulation and Characterization of the Double Network Hydrogel Surface-Bulk Transition. Macromolecules, 52, 6704-6713, 2019.
- Huang J, Frauenlob M, Shibata Y, Wang L, Nakajima T, Nonoyama T, Tsuda M, Tanaka S, Kurokawa T, and Gong J P. Chitin-Based Double-Network Hydrogel as Potential Superficial Soft-Tissue-Repairing Materials. Biomacromolecules, 21, 4220-4230, 2020.
- Nonoyama T, Wang L, Tsuda M, Suzuki Y, Kiyama R, Yasuda K, Tanaka S, Nagata K, Fujita R, Sakamoto N, Kawasaki N, Yurimoto H, and Gong J P. Isotope Microscopic Observation of Osteogenesis Process Forming Robust Bonding of Double Network Hydrogel to Bone. Adv Healthc Mater, e2001731, 2020.
- Kaibara T, Wang L, Tsuda M, Nonoyama T, Kurokawa T, Iwasaki N, Gong J P, Tanaka S, and Yasuda K. Hydroxyapatite-hybridized double-network hydrogel surface enhances differentiation of bone marrow-derived mesenchymal stem cells to osteogenic cells. J Biomed Mater Res A, 2021.
- Mu Q, Cui K, Wang Z J, Matsuda T, Cui W, Kato H, Namiki S, Yamazaki T, Frauenlob M, Nonoyama T, Tsuda M, Tanaka S, Nakajima T, and Gong J P. Force-triggered rapid microstructure growth on hydrogel surface for on-demand functions. Nat Commun, 13, 6213, 2022.
Ⅱ. Research for SARS-CoV-2
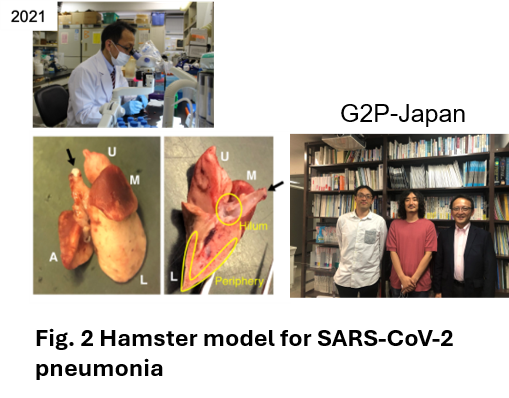
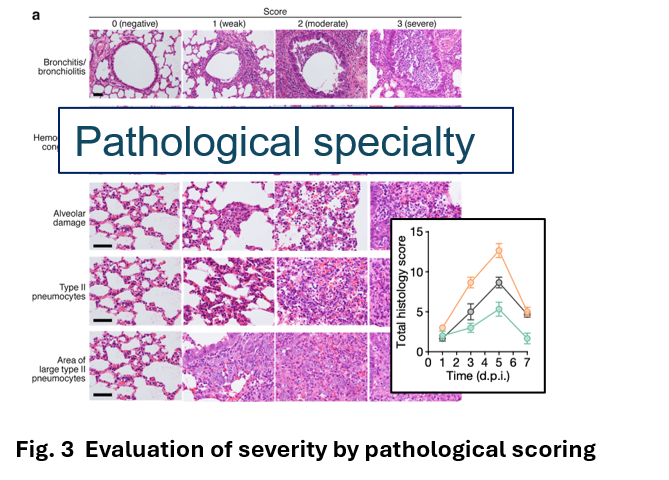
In 2021 with coronavirus disaster, Dr. Fukuhara (Hokkaido University Department of Microbiology) asked me to look at pneumonia of hamsters infected with coronavirus. Although Dr. Tanaka had no experience in analyzing hamsters, He first prepared specimens by cutting them out (Fig. 2) and established a scoring method for tissue evaluation by reviewing H&E specimens (Fig. 3) from early in the morning to late at night during the week of Japanese traditional Bon festival time(2). When the research paper on the SARS-CoV-2 delta strain was accepted in Nature (Fig. 4)and this histopathological analysis seemed to be useful. Then, Omicron emerged and pathogenicity became a social issue. The virus arrived at Hokkaido University on December 10, and Tanaka and his colleagues worked on the analysis for two weeks without a sleep time to get the results. Dr. Sato (Univ. of Tokyo) who is the chief scientist of G2P-Japan mentioned the results via Twitter on December 26, and the next day, the results were reported on the TV news show as News Station (TBS). Dr. Tanaka says ‘although I am neither a virologist nor a respiratory pathologist myself, I found that the reason why I could make-up histological evaluation system of coronavirus infection mostly depends on my carrier as pathologist always discussing with the respiratory physicians at the daily clinic-pathological conferences (CPCs). I realized once again that when an unknown pathogen appears histopathological analysis by pathology experts is extremely important to elucidate the pathogenesis. I wish to share this information with young pathologists and researchers.’
Publications for Research for SARS-CoV-2
- Kimura I, Yamasoba D, Tamura T, Nao N, Suzuki T, Oda Y, Mitoma S, Ito J, Nasser H, Zahradnik J, Uriu K, Fujita S, Kosugi Y, Wang L, Tsuda M, Kishimoto M, Ito H, Suzuki R, Shimizu R, Begum M M, Yoshimatsu K, Kimura K T, Sasaki J, Sasaki-Tabata K, Yamamoto Y, Nagamoto T, Kanamune J, Kobiyama K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Shirakawa K, Takaori-Kondo A, Kuramochi J, Schreiber G, Ishii K J, Hashiguchi T, Ikeda T, Saito A, Fukuhara T, Tanaka S, Matsuno K, and Sato K. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell, 185, 3992-4007.e3916, 2022.
- Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, Kosugi Y, Shirakawa K, Sadamasu K, Kimura I, Ito J, Wu J, Iwatsuki-Horimoto K, Ito M, Yamayoshi S, Loeber S, Tsuda M, Wang L, Ozono S, Butlertanaka E P, Tanaka Y L, Shimizu R, Shimizu K, Yoshimatsu K, Kawabata R, Sakaguchi T, Tokunaga K, Yoshida I, Asakura H, Nagashima M, Kazuma Y, Nomura R, Horisawa Y, Yoshimura K, Takaori-Kondo A, Imai M, Chiba M, Furihata H, Hasebe H, Kitazato K, Kubo H, Misawa N, Morizako N, Noda K, Oide A, Suganami M, Takahashi M, Tsushima K, Yokoyama M, Yuan Y, Tanaka S, Nakagawa S, Ikeda T, Fukuhara T, Kawaoka Y, Sato K, and The Genotype to Phenotype Japan C. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature, 602, 300-306, 2022.
- Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, Kosugi Y, Tsuda M, Orba Y, Sasaki M, Shimizu R, Kawabata R, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Sawa H, Ikeda T, Irie T, Matsuno K, Tanaka S, Fukuhara T, and Sato K. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature, 603, 700-705, 2022.
- Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, Uriu K, Tsuda M, Zahradnik J, Shirakawa K, Suzuki R, Kishimoto M, Kosugi Y, Kobiyama K, Hara T, Toyoda M, Tanaka Y L, Butlertanaka E P, Shimizu R, Ito H, Wang L, Oda Y, Orba Y, Sasaki M, Nagata K, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Kuramochi J, Seki M, Fujiki R, Kaneda A, Shimada T, Nakada T A, Sakao S, Suzuki T, Ueno T, Takaori-Kondo A, Ishii K J, Schreiber G, Sawa H, Saito A, Irie T, Tanaka S, Matsuno K, Fukuhara T, Ikeda T, and Sato K. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell, 185, 2103-2115.e2119, 2022.
- Ito J, Suzuki R, Uriu K, Itakura Y, Zahradnik J, Kimura K T, Deguchi S, Wang L, Lytras S, Tamura T, Kida I, Nasser H, Shofa M, Begum M M, Tsuda M, Oda Y, Suzuki T, Sasaki J, Sasaki-Tabata K, Fujita S, Yoshimatsu K, Ito H, Nao N, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Yamamoto Y, Nagamoto T, Kuramochi J, Schreiber G, Saito A, Matsuno K, Takayama K, Hashiguchi T, Tanaka S, Fukuhara T, Ikeda T, and Sato K. Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant. Nat Commun, 14, 2671, 2023.
- Tamura T, Ito J, Uriu K, Zahradnik J, Kida I, Anraku Y, Nasser H, Shofa M, Oda Y, Lytras S, Nao N, Itakura Y, Deguchi S, Suzuki R, Wang L, Begum M M, Kita S, Yajima H, Sasaki J, Sasaki-Tabata K, Shimizu R, Tsuda M, Kosugi Y, Fujita S, Pan L, Sauter D, Yoshimatsu K, Suzuki S, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Yamamoto Y, Nagamoto T, Schreiber G, Maenaka K, Hashiguchi T, Ikeda T, Fukuhara T, Saito A, Tanaka S, Matsuno K, Takayama K, and Sato K. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat Commun, 14, 2800, 2023.
- Tamura T, Yamasoba D, Oda Y, Ito J, Kamasaki T, Nao N, Hashimoto R, Fujioka Y, Suzuki R, Wang L, Ito H, Kashima Y, Kimura I, Kishimoto M, Tsuda M, Sawa H, Yoshimatsu K, Yamamoto Y, Nagamoto T, Kanamune J, Suzuki Y, Ohba Y, Yokota I, Matsuno K, Takayama K, Tanaka S, Sato K, and Fukuhara T. Comparative pathogenicity of SARS-CoV-2 Omicron subvariants including BA.1, BA.2, and BA.5. Commun Biol, 6, 772, 2023.
- Fujita S, Plianchaisuk A, Deguchi S, Ito H, Nao N, Wang L, Nasser H, Tamura T, Kimura I, Kashima Y, Suzuki R, Suzuki S, Kida I, Tsuda M, Oda Y, Hashimoto R, Watanabe Y, Uriu K, Yamasoba D, Guo Z, Hinay A A, Jr., Kosugi Y, Chen L, Pan L, Kaku Y, Chu H, Donati F, Temmam S, Eloit M, Yamamoto Y, Nagamoto T, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Suzuki Y, Ito J, Ikeda T, Tanaka S, Matsuno K, Fukuhara T, Takayama K, and Sato K. Virological characteristics of a SARS-CoV-2-related bat coronavirus, BANAL-20-236. EBioMedicine, 104, 105181, 2024.
- Ito H, Tamura T, Wang L, Mori K, Tsuda M, Suzuki R, Suzuki S, Yoshimatsu K, Tanaka S, and Fukuhara T. Involvement of SARS-CoV-2 accessory proteins in immunopathogenesis. Microbiol Immunol, 68, 237-247, 2024.
- Tamura T, Ito H, Torii S, Wang L, Suzuki R, Tsujino S, Kamiyama A, Oda Y, Tsuda M, Morioka Y, Suzuki S, Shirakawa K, Sato K, Yoshimatsu K, Matsuura Y, Iwano S, Tanaka S, and Fukuhara T. Akaluc bioluminescence offers superior sensitivity to track in vivo dynamics of SARS-CoV-2 infection. iScience, 27, 109647, 2024.
- Tamura T, Mizuma K, Nasser H, Deguchi S, Padilla-Blanco M, Oda Y, Uriu K, Tolentino J E M, Tsujino S, Suzuki R, Kojima I, Nao N, Shimizu R, Wang L, Tsuda M, Jonathan M, Kosugi Y, Guo Z, Hinay A A, Jr., Putri O, Kim Y, Tanaka Y L, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Saito A, Ito J, Irie T, Tanaka S, Zahradnik J, Ikeda T, Takayama K, Matsuno K, Fukuhara T, and Sato K. Virological characteristics of the SARS-CoV-2 BA.2.86 variant. Cell Host Microbe, 2024.
- Tsujino S, Deguchi S, Nomai T, Padilla-Blanco M, Plianchaisuk A, Wang L, Begum M M, Uriu K, Mizuma K, Nao N, Kojima I, Tsubo T, Li J, Matsumura Y, Nagao M, Oda Y, Tsuda M, Anraku Y, Kita S, Yajima H, Sasaki-Tabata K, Guo Z, Hinay A A, Jr., Yoshimatsu K, Yamamoto Y, Nagamoto T, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Nasser H, Jonathan M, Putri O, Kim Y, Chen L, Suzuki R, Tamura T, Maenaka K, Irie T, Matsuno K, Tanaka S, Ito J, Ikeda T, Takayama K, Zahradnik J, Hashiguchi T, Fukuhara T, and Sato K. Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant. Microbiol Immunol, 2024.
Ⅲ. Basic cancer research: Oncogene and molecular mechanism of cancer progression
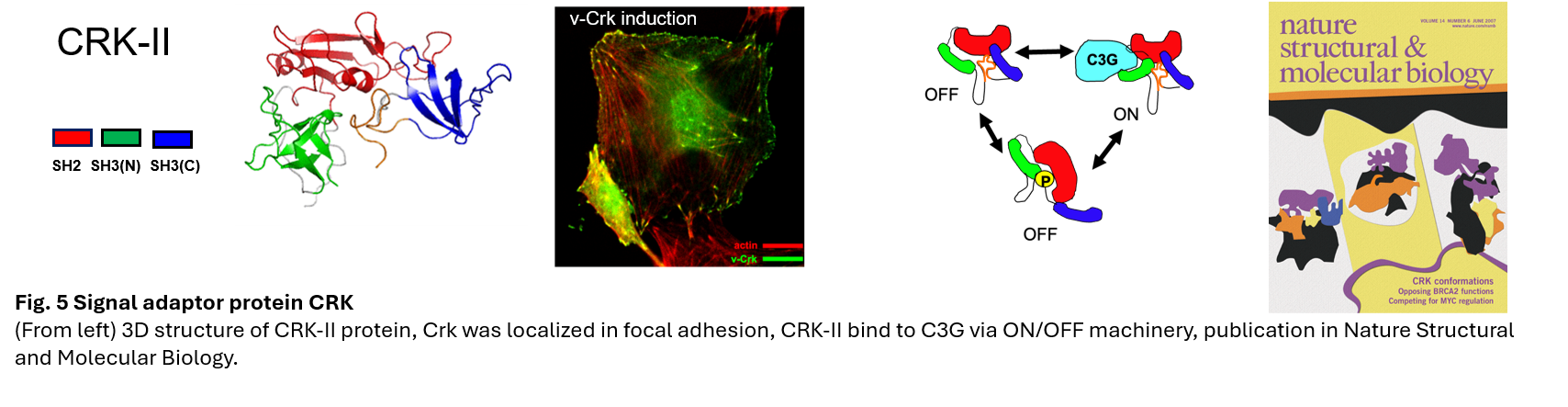
Dr. Tanaka studied CRK, an adaptor molecule for intracellular signal transduction, under Dr. Michiyuki Matsuda (Professor emeritus of Kyoto University) in the 1990s, and cloned the novel genes C3G and DOCK180, which are low molecular weight G protein activators (GEFs), and discovered a tyrosine kinase → adaptor molecule→GEF→Ras→ERK signaling pathway (Tanaka, Mol Cell Biol, 1993; Tanaka, Proc Natl Acad Sci, 1994). This is now a major pathway in molecular targeted cancer therapy. Dr. Tanaka then studied in the laboratory of Dr. Hidesaburo Hanafusa (Lasker Award winner) at Rockefeller University in the U.S., where he worked to elucidate the mechanism of oncogenesis. After returning to Japan, Dr. Tanaka clarified the three-dimensional structure of CRK for the development of therapeutic drugs (Kobashigawa, Nat Struct Mol Biol, 2007). In the field of rare cancer research, which is now a social issue, he identified the SS18-SSX fusion gene as the cause of synovial sarcoma (Nagai, Proc Natl Acad Sci , 2001). In cancer stem cell research, he isolated the CD133 promoter region of brain tumor stem cells and elucidated the mechanism of drug resistance (Tabu, Cell Res, 2008) Requests for the distribution of the CD133 promoter have come from all over the world, and it is provided free of charge. He also elucidated that sarcoma stem cells are CXCR4 positive (Kimura, Oncogene, 2015).
Publications for Basic cancer research: Oncogene and molecular mechanism of cancer progression
- Tanaka, S., Morishita, T., Hashimoto, Y., Hattori, S., Nakamura, S., Shibuya, M., Matuoka, K., Takenawa, T., Kurata, T., Nagashima, K., and et al. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci U S A, 91, 3443-3447, 1994.
- Tanaka, S., Ouchi, T., and Hanafusa, H. Downstream of Crk adaptor signaling pathway: activation of Jun kinase by v-Crk through the guanine nucleotide exchange protein C3G. Proc Natl Acad Sci U S A, 94, 2356-2361, 1997.
- Nagai, M., Tanaka, S., Tsuda, M., Endo, S., Kato, H., Sonobe, H., Minami, A., Hiraga, H., Nishihara, H., Sawa, H., and Nagashima, K. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci U S A, 98, 3843-3848, 2001.
- Tsuda, M., Makino, Y., Iwahara, T., Nishihara, H., Sawa, H., Nagashima, K., Hanafusa, H., and Tanaka, S. Crk associates with ERM proteins and promotes cell motility toward hyaluronic acid. J Biol Chem, 279, 46843-46850, 2004.
- Kobashigawa, Y., Sakai, M., Naito, M., Yokochi, M., Kumeta, H., Makino, Y., Ogura, K., Tanaka, S., and Inagaki, F. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat Struct Mol Biol, 14, 503-510, 2007.
- Kohsaka, S., Wang, L., Yachi, K., Mahabir, R., Narita, T., Itoh, T., Tanino, M., Kimura, T., Nishihara, H., and Tanaka, S. STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol Cancer Ther, 11, 1289-1299, 2012.
- Kimura, T., Wang, L., Tabu, K., Tsuda, M., Tanino, M., Maekawa, A., Nishihara, H., Hiraga, H., Taga, T., Oda, Y., and Tanaka, S. Identification and analysis of CXCR4-positive synovial sarcoma-initiating cells. Oncogene, 35, 3932-3943, 2016.
- Matsumoto, R., Tsuda, M., Yoshida, K., Tanino, M., Kimura, T., Nishihara, H., Abe, T., Shinohara, N., Nonomura, K., and Tanaka, S. Aldo-keto reductase 1C1 induced by interleukin-1beta mediates the invasive potential and drug resistance of metastatic bladder cancer cells. Sci Rep, 6, 34625, 2016.
- Yachi, K., Tsuda, M., Kohsaka, S., Wang, L., Oda, Y., Tanikawa, S., Ohba, Y., and Tanaka, S. miR-23a promotes invasion of glioblastoma via HOXD10-regulated glial-mesenchymal transition. Signal Transduction and Targeted Therapy, 3, 33, 2018.
- Tsuda, M., Noguchi, M., Kurai, T., Ichihashi, Y., Ise, K., Wang, L., Ishida, Y., Tanino, M., Hirano, S., Asaka, M., and Tanaka, S. Aberrant expression of MYD88 via RNA-controlling CNOT4 and EXOSC3 in colonic mucosa impacts generation of colonic cancer. Cancer Sci, 112, 5100-5113, 2021.
- Tsuda, M., Horio, R., Wang, L., Takenami, T., Moriya, J., Suzuka, J., Sugino, H., Tanei, Z., Tanino, M., and Tanaka, S. Novel rapid immunohistochemistry using an alternating current electric field identifies Rac and Cdc42 activation in human colon cancer FFPE tissues. Sci Rep, 12, 1733, 2022.
- Zhai, T., Mitamura, T., Wang, L., Kubota, S.I., Murakami, M., Tanaka, S., and Watari, H. Combination therapy with bevacizumab and a CCR2 inhibitor for human ovarian cancer: An in vivo validation study. Cancer Med, 12, 9697-9708, 2023.
- Hirose, T., Ikegami, M., Kida, K., Ueno, T., Kitada, R., Wang, L., Tanaka, S., Endo, M., Nakashima, Y., Kanomata, N., Mano, H., Yamauchi, H., and Kohsaka, S. Cancer risk assessment of premalignant breast tissues from patients with BRCA mutations by genome profiling. NPJ Breast Cancer, 10, 87, 2024.
- Sun, X., Watanabe, T., Oda, Y., Shen, W., Ahmad, A., Ouda, R., de Figueiredo, P., Kitamura, H., Tanaka, S., and Kobayashi, K.S. Targeted demethylation and activation of NLRC5 augment cancer immunogenicity through MHC class I. Proc Natl Acad Sci USA, 121, e2310821121, 2024.
- Wang, M., Kono, M., Yamaguchi, Y., Islam, J., Shoji, S., Kitagawa, Y., Fushimi, K., Watanabe, S., Matsuba, G., Yamamoto, A., Tanaka, M., Tsuda, M., Tanaka, S., and Hasegawa, Y. Structure-changeable luminescent Eu(III) complex as a human cancer grade probing system for brain tumor diagnosis. Sci Rep, 14, 778, 2024.
IV. Brain tumor research
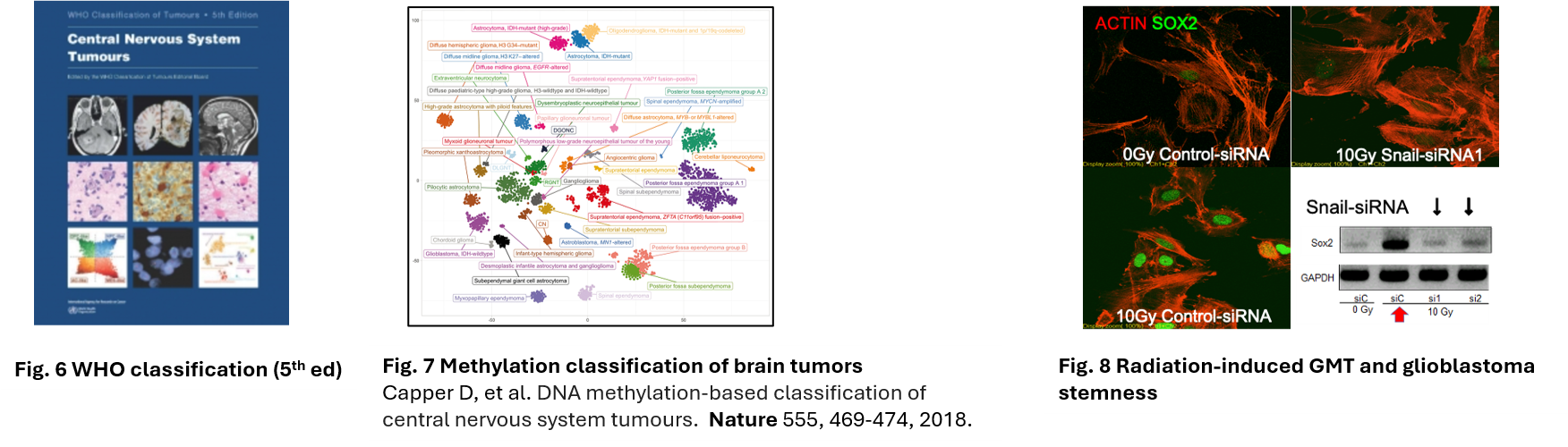
As a pathologist, Dr. Tanaka’s subspecialty is neuropathology, especially for the diagnosis of brain tumor. He is the one of the authors of WHO tumour classification of central nervous system (5th ed.)(Fig. 6). Thus, Dr. Tanaka has bee studying in the molecular mechanism of malignant brain tumor including glioblastoma. Glioblastoma is a poor prognosis tumor with a 5-year survival rate of less than 10%, most often in the elderly, but it appears as a ring-enhancing mass on imaging, and histology shows fenestrated necrosis, multinucleated giant cells and microangiogenesis, currently defined as IDH wild type glioblastoma in the WHO 5th edition. IDH mutations produce 2HG oncometabolite, which regulates methyltransferases and induces a methylated phenotype called CIMP. The WHO is currently in its 5th edition, but the brain tumor classification was based on the “histological diagnosis based on lineage development” by Bailey-Cushing et al. in 1926, but now it is based on methylation analysis
(Nature 555, 469-474, 2018)(Fig. 7). Glioblastoma IDH wild type is considered to be a glioblastoma caused by the accumulation of genetic mutations in the progenitor cells.
Initially, Tanaka lab worked on overcoming chemotherapy resistance in glioblastoma. Temozolomide is still used as a standard treatment, but because it is an alkylating agent, DNA methylation is the drug‘s effect, and high levels of MGMT, a methyltransferase enzyme, cancel the drug’s effect. He first proposed a method for evaluating MGMT using IHC. Then, Dr. Tanaka focused on the mesenchymal type of glioblastoma, which has a poor prognosis, with the aim of developing a drug that suppresses MGMT. Although regulatory factors were reported for the mesenchymal type, using pathological tissues, we found that the amount of phosphorylated STAT3 positively correlated with the amount of MGMT. The Tanaka team then found that the expression of MGMT was decreased by jack inhibitors. They found that the drug effect of temozolomide was enhanced by the combination of this inhibitor. Next, since radiation is one of the standard treatments for glioblastoma, we worked to elucidate its resistance
mechanism. Histopathology revealed that Vimentin expression is increased in recurrence after radiotherapy of glioblastoma, and in fact irradiation of cells enhances cell adhesion plaques. Irradiation was found to increase Snail and induce mesenchymal features via activation of the ERK pathway, and to enhance the production of TGF-beta from surrounding macrophages and astrocytes, which led to the concept of GMT, glia- mesenchumal transition. They also found that SOX expression is increased during this process, which may induce glioblastoma stemness (Fig. 8). Therefore, Dr. Tanaka established the concept for glial-mesenchymal transition (GMT) with GBM stemness.
Publications for Brain tumor research
- Sasai, K., Akagi, T., Aoyanagi, E., Tabu, K., Kaneko, S., and Tanaka, S. O6-methylguanine-DNA methyltransferase is downregulated in transformed astrocyte cells: implications for anti-glioma therapies. Mol Cancer, 6, 36, 2007.
- Tabu, K., Ohba, Y., Suzuki, T., Makino, Y., Kimura, T., Ohnishi, A., Sakai, M., Watanabe, T., Tanaka, S., and Sawa, H. Oligodendrocyte lineage transcription factor 2 inhibits the motility of a human glial tumor cell line by activating RhoA. Mol Cancer Res, 5, 1099-1109, 2007.
- Wang, L., Tabu, K., Kimura, T., Tsuda, M., Linghu, H., Tanino, M., Kaneko, S., Nishihara, H., and Tanaka, S. Signaling adaptor protein Crk is indispensable for malignant feature of glioblastoma cell line KMG4. Biochem Biophys Res Commun, 362, 976-981, 2007.
- Tabu, K., Sasai, K., Kimura, T., Wang, L., Aoyanagi, E., Kohsaka, S., Tanino, M., Nishihara, H., and Tanaka, S. Promoter hypomethylation regulates CD133 expression in human gliomas. Cell Res, 18, 1037-1046, 2008.
- Wang, L., Sasai, K., Akagi, T., and Tanaka, S. Establishment of a luciferase assay-based screening system: fumitremorgin C selectively inhibits cellular proliferation of immortalized astrocytes expressing an active form of AKT. Biochem Biophys Res Commun, 373, 392-396, 2008.
- Kanno, H., Nishihara, H., Narita, T., Yamaguchi, S., Kobayashi, H., Tanino, M., Kimura, T., Terasaka, S., and Tanaka, S. Prognostic implication of histological oligodendroglial tumor component: clinicopathological analysis of 111 cases of malignant gliomas. PLoS One, 7, e41669, 2012.
- Kohsaka, S., Wang, L., Yachi, K., Mahabir, R., Narita, T., Itoh, T., Tanino, M., Kimura, T., Nishihara, H., and Tanaka, S. STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol Cancer Ther, 11, 1289-1299, 2012.
- Kanno, H., Nishihara, H., Wang, L., Yuzawa, S., Kobayashi, H., Tsuda, M., Kimura, T., Tanino, M., Terasaka, S., and Tanaka, S. Expression of CD163 prevents apoptosis through the production of granulocyte colony-stimulating factor in meningioma. Neuro Oncol, 15, 853-864, 2013.
- Mahabir, R., Tanino, M., Elmansuri, A., Wang, L., Kimura, T., Itoh, T., Ohba, Y., Nishihara, H., Shirato, H., Tsuda, M., and Tanaka, S. Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma. Neuro Oncol, 16, 671-685, 2014.
- Ishi Y, Takamiya S, Seki T, Yamazaki K, Hida K, Hatanaka K C, Ishida Y, Oda Y, Tanaka S, and Yamaguchi S. Prognostic role of H3K27M mutation, histone H3K27 methylation status, and EZH2 expression in diffuse spinal cord gliomas. Brain Tumor Pathol, 37, 81-88, 2020.
- Habiba U, Sugino H, Yordanova R, Ise K, Tanei Z I, Ishida Y, Tanikawa S, Terasaka S, Sato K I, Kamoshima Y, Katoh M, Nagane M, Shibahara J, Tsuda M, and Tanaka S. Loss of H3K27 trimethylation is frequent in IDH1-R132H but not in non-canonical IDH1/2 mutated and 1p/19q codeleted oligodendroglioma: a Japanese cohort study. Acta Neuropathol Commun, 9, 95, 2021.
V. Surgical pathology
Dr. Tanaka is professor of Deparment of Cancer Pathology, thus various projects of surgical pathology research are on going. Especially, several important papers for establishment of criteria of surgical pathological diagnosis have been published in Tanaka lab including AJSP, Mod Path, Gastroenteology, and so on.
Publications for Surgical pathology
- Nishihara H, Tanaka S, Tsuda M, Oikawa S, Maeda M, Shimizu M, Shinomiya H, Tanigami A, Sawa H, and Nagashima K. Molecular and immunohistochemical analysis of signaling adaptor protein Crk in human cancers. Cancer Lett, 180, 55-61, 2002.
- Orba Y, Tanaka S, Nishihara H, Kawamura N, Itoh T, Shimizu M, Sawa H, and Nagashima K. Application of laser capture microdissection to cytologic specimens for the detection of immunoglobulin heavy chain gene rearrangement in patients with malignant lymphoma. Cancer, 99, 198-204, 2003.
- Sasai K, Nodagashira M, Nishihara H, Aoyanagi E, Wang L, Katoh M, Murata J, Ozaki Y, Ito T, Fujimoto S, Kaneko S, Nagashima K, and Tanaka S. Careful exclusion of non-neoplastic brain components is required for an appropriate evaluation of O6-methylguanine-DNA methyltransferase status in glioma: relationship between immunohistochemistry and methylation analysis. Am J Surg Pathol, 32, 1220-1227, 2008.
- Kanno H, Nishihara H, Oikawa M, Ozaki Y, Murata J, Sawamura Y, Kato M, Kubota K, Tanino M, Kimura T, Nagashima K, Itoh T, and Tanaka S. Expression of O(6)-methylguanine DNA methyltransferase (MGMT) and immunohistochemical analysis of 12 pineal parenchymal tumors. Neuropathology, 32, 647-653, 2012.
- Yuzawa S, Kano M R, Einama T, and Nishihara H. PDGFRbeta expression in tumor stroma of pancreatic adenocarcinoma as a reliable prognostic marker. Med Oncol, 29, 2824-2830, 2012.
- Tanino M, Sasajima T, Nanjo H, Akesaka S, Kagaya M, Kimura T, Ishida Y, Oda M, Takahashi M, Sugawara T, Yoshioka T, Nishihara H, Akagami Y, Goto A, Minamiya Y, Tanaka S, and Group R I S. Rapid immunohistochemistry based on alternating current electric field for intraoperative diagnosis of brain tumors. Brain Tumor Pathol, 32, 12-19, 2014.
- Moriya J, Tanino M A, Takenami T, Endoh T, Urushido M, Kato Y, Wang L, Kimura T, Tsuda M, Nishihara H, Tanaka S, and Group R I S. Rapid immunocytochemistry based on alternating current electric field using squash smear preparation of central nervous system tumors. Brain Tumor Pathol, 33, 13-18, 2016.
- Yuzawa S, Nishihara H, Wang L, Tsuda M, Kimura T, Tanino M, and Tanaka S. Analysis of NAB2-STAT6 Gene Fusion in 17 Cases of Meningeal Solitary Fibrous Tumor/Hemangiopericytoma: Review of the Literature. Am J Surg Pathol, 40, 1031-1041, 2016.
- Yuzawa S, Nishihara H, Yamaguchi S, Mohri H, Wang L, Kimura T, Tsuda M, Tanino M, Kobayashi H, Terasaka S, Houkin K, Sato N, and Tanaka S. Clinical impact of targeted amplicon sequencing for meningioma as a practical clinical-sequencing system. Mod Pathol, 29, 708-716, 2016.
- Hirose T, Nobusawa S, Sugiyama K, Amatya V J, Fujimoto N, Sasaki A, Mikami Y, Kakita A, Tanaka S, and Yokoo H. Astroblastoma: A Distinct Tumor Entity Characterized by Alterations of the X Chromosome and MN1 Rearrangement. Brain Pathol, 2017.
- Omori Y, Ono Y, Tanino M, Karasaki H, Yamaguchi H, Furukawa T, Enomoto K, Ueda J, Sumi A, Katayama J, Muraki M, Taniue K, Takahashi K, Ambo Y, Shinohara T, Nishihara H, Sasajima J, Maguchi H, Mizukami Y, Okumura T, and Tanaka S. Pathways of Progression From Intraductal Papillary Mucinous Neoplasm to Pancreatic Ductal Adenocarcinoma Based on Molecular Features. Gastroenterology, 156, 647-661.e642, 2019.
- Takayama Y, Ono Y, Mizukami Y, Itoh H, Nakajima N, Arai H, Tanaka S, Nobusawa S, Yokoo H, and Onozato Y. Comparative genome-wide analysis of gastric adenocarcinomas with hyperplastic polyp components. Virchows Arch, 475, 383-389, 2019.
- Tanei Z I, Saito Y, Ito S, Matsubara T, Motoda A, Yamazaki M, Sakashita Y, Kawakami I, Ikemura M, Tanaka S, Sengoku R, Arai T, and Murayama S. Lewy pathology of the esophagus correlates with the progression of Lewy body disease: a Japanese cohort study of autopsy cases. Acta Neuropathol, 141, 25-37, 2021.
- Shirakura T, Yamada Y, Nakata S, Asayama B, Seo Y, Tanikawa S, Kato T, Komoribayashi N, Kubo N, Monma N, Okura N, Tanaka S, Oda Y, Hirato J, Yokoo H, and Nobusawa S. Analysis of clinicopathological features and NAB2-STAT6 fusion variants of meningeal solitary fibrous tumor with ectopic salivary gland components in the cerebellopontine angle. Virchows Arch, 2022.
VI. AI and digital pathology
Dr. Tanaka extensively proceeds the research on digital pathology and AI-based medical researches
Publications for AI and digital pathology
- Takiyama A, Teramoto T, Suzuki H, Yamashiro K, and Tanaka S. Persistent homology index as a robust quantitative measure of immunohistochemical scoring. Sci Rep, 7, 14002, 2017.
- Shimizu H, Enda K, Koyano H, Ogawa T, Takahashi D, Tanaka S, Iwasaki N, and Shimizu T. Diagnosis on Ultrasound Images for Developmental Dysplasia of the Hip with a Deep Learning-Based Model Focusing on Signal Heterogeneity in the Bone Region. Diagnostics (Basel), 15, 2025.
- Oshino T, Enda K, Shimizu H, Sato M, Nishida M, Kato F, Oda Y, Hosoda M, Kudo K, Iwasaki N, Tanaka S, and Takahashi M. Artificial intelligence can extract important features for diagnosing axillary lymph node metastasis in early breast cancer using contrast-enhanced ultrasonography. Sci Rep, 15, 5648, 2025.
- Shimizu H, Enda K, Koyano H, Shimizu T, Shimodan S, Sato K, Ogawa T, Tanaka S, Iwasaki N, and Takahashi D. Bimodal machine learning model for unstable hips in infants: integration of radiographic images with automatically-generated clinical measurements. Sci Rep, 14, 17826, 2024.
VII. Other collaborative researches
Dr. Tanaka extensively proceeds the collaboration study with various fields of medical sciences.
Publications for Other collaborative researches
- Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, Arase N, Shiratori I, Tanaka S, Kawaguchi Y, Spear P G, Lanier L L, and Arase H. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell, 132, 935-944, 2008.
- Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, Higashi H, Musashi M, Iwabuchi K, Suzuki M, Yamada G, Azuma T, and Hatakeyama M. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A, 105, 1003-1008, 2008.
- Shime H, Matsumoto M, Oshiumi H, Tanaka S, Nakane A, Iwakura Y, Tahara H, Inoue N, and Seya T. Toll-like receptor 3 signaling converts tumor-supporting myeloid cells to tumoricidal effectors. Proc Natl Acad Sci U S A, 109, 2066-2071, 2012.
- Michels S, Trautmann M, Sievers E, Kindler D, Huss S, Renner M, Friedrichs N, Kirfel J, Steiner S, Endl E, Wurst P, Heukamp L, Penzel R, Larsson O, Kawai A, Tanaka S, Sonobe H, Schirmacher P, Mechtersheimer G, Wardelmann E, Buttner R, and Hartmann W. SRC signaling is crucial in the growth of synovial sarcoma cells. Cancer Res, 73, 2518-2528, 2013.
- Li S, Zhang P, Freibaum B D, Kim N C, Kolaitis R M, Molliex A, Kanagaraj A P, Yabe I, Tanino M, Tanaka S, Sasaki H, Ross E D, Taylor J P, and Kim H J. Genetic interaction of hnRNPA2B1 and DNAJB6 in a Drosophila model of multisystem proteinopathy. Hum Mol Genet, 25, 936-950, 2016.
- Kawano S, Grassian A R, Tsuda M, Knutson S K, Warholic N M, Kuznetsov G, Xu S, Xiao Y, Pollock R M, Smith J S, Kuntz K K, Ribich S, Minoshima Y, Matsui J, Copeland R A, Tanaka S, and Keilhack H. Preclinical Evidence of Anti-Tumor Activity Induced by EZH2 Inhibition in Human Models of Synovial Sarcoma. PLoS One, 11, e0158888, 2016.
- Yabe I, Yaguchi H, Kato Y, Miki Y, Takahashi H, Tanikawa S, Shirai S, Takahashi I, Kimura M, Hama Y, Matsushima M, Fujioka S, Kano T, Watanabe M, Nakagawa S, Kunieda Y, Ikeda Y, Hasegawa M, Nishihara H, Ohtsuka T, Tanaka S, Tsuboi Y, Hatakeyama S, Wakabayashi K, and Sasaki H. Mutations in bassoon in individuals with familial and sporadic progressive supranuclear palsy-like syndrome. Sci Rep, 8, 819, 2018.
- Sun X, Watanabe T, Oda Y, Shen W, Ahmad A, Ouda R, de Figueiredo P, Kitamura H, Tanaka S, and Kobayashi K S. Targeted demethylation and activation of NLRC5 augment cancer immunogenicity through MHC class I. Proc Natl Acad Sci U S A, 121, e2310821121, 2024.
